- 1Animal Breeding and Genomics, Wageningen University and Research, Wageningen, Netherlands
- 2Farm Technology, Wageningen University and Research, Wageningen, Netherlands
- 3Animal Health and Welfare, Wageningen University and Research, Wageningen, Netherlands
Animal pose-estimation networks enable automated estimation of key body points in images or videos. This enables animal breeders to collect pose information repeatedly on a large number of animals. However, the success of pose-estimation networks depends in part on the availability of data to learn the representation of key body points. Especially with animals, data collection is not always easy, and data annotation is laborious and time-consuming. The available data is therefore often limited, but data from other species might be useful, either by itself or in combination with the target species. In this study, the across-species performance of animal pose-estimation networks and the performance of an animal pose-estimation network trained on multi-species data (turkeys and broilers) were investigated. Broilers and turkeys were video recorded during a walkway test representative of the situation in practice. Two single-species and one multi-species model were trained by using DeepLabCut and tested on two single-species test sets. Overall, the within-species models outperformed the multi-species model, and the models applied across species, as shown by a lower raw pixel error, normalized pixel error, and higher percentage of keypoints remaining (PKR). The multi-species model had slightly higher errors with a lower PKR than the within-species models but had less than half the number of annotated frames available from each species. Compared to the single-species broiler model, the multi-species model achieved lower errors for the head, left foot, and right knee keypoints, although with a lower PKR. Across species, keypoint predictions resulted in high errors and low to moderate PKRs and are unlikely to be of direct use for pose and gait assessments. A multi-species model may reduce annotation needs without a large impact on performance for pose assessment, however, with the recommendation to only be used if the species are comparable. If a single-species model exists it could be used as a pre-trained model for training a new model, and possibly require a limited amount of new data. Future studies should investigate the accuracy needed for pose and gait assessments and estimate genetic parameters for the new phenotypes before pose-estimation networks can be applied in practice.
Introduction
In poultry production, locomotion is an important health and welfare trait. Impaired locomotion is a major welfare concern (Scientific Committee on Animal Health and Animal Welfare, 2000; van Staaveren et al., 2020), and a cause of economic losses in both turkeys and broilers (Sullivan, 1994; van Staaveren et al., 2020). Impaired locomotion has been linked to high growth rate, high body weight, infection, and housing conditions (e.g., light and feeding regime) in broilers (Bradshaw et al., 2002). Birds with impaired locomotion have trouble accessing feed and water (Weeks et al., 2000), performing motivated behaviors like dust bathing (Vestergaard and Sanotra, 1999), and likely with peck avoidance (Erasmus, 2018). Studies have reported that in broilers approximately 15–28% of the birds, and in turkeys, approximately 8–13% of the birds, examined had impaired locomotion (Kestin et al., 1992; Bassler et al., 2013; Sharafeldin et al., 2015; Vermette et al., 2016; Kittelsen et al., 2017).
Gait-scoring systems have been developed for both turkeys and broilers (e.g., Kestin et al., 1992; Garner et al., 2002; Quinton et al., 2011; Kapell et al., 2017). Generally, a human expert judges the gait of an animal from behind, or the side, based on several locomotion factors, which often relate to the fluidity of movement and leg conformation. Gait scores were found to be heritable in turkeys [h2: 0.08–0.13 ± 0.01 (Kapell et al., 2017) and 0.25–0.26 ± 0.01 (Quinton et al., 2011)]. The gait scores are valuable to breeding programs, yet the gait-scoring process is laborious, and gait scores are prone to subjectivity. Sensor technologies could provide relatively effortless, non-invasive, and objective gait assessments, while also allowing for the assessment of a larger number of animals with higher frequency.
Several technologies for objective gait assessment have been introduced over the years. These technologies include pressure-sensitive walkways (PSW) (Nääs et al., 2010; Paxton et al., 2013; Oviedo-Rondón et al., 2017; Kremer et al., 2018; Stevenson et al., 2019), rotarods (Malchow et al., 2019), video analysis (Abourachid, 1991, 2001; Caplen et al., 2012; Paxton et al., 2013; Oviedo-Rondón et al., 2017), accelerometers (Stevenson et al., 2019), and inertial measurement units (IMUs; provide 3D accelerometer, gyroscope, and, occasionally, magnetometer data) (Bouwman et al., 2020). The on-farm application of the sensor technologies is limited due to equipment costs (PSW), habituation requirements (PSW, accelerometers, IMUs), or increased animal handling (rotarod, accelerometers, and IMUs). On-farm application of cameras can be more practical, however, the investigated camera-based methods rely on physical markers placed on key body points to assess the gait of an animal (Caplen et al., 2012; Paxton et al., 2013; Oviedo-Rondón et al., 2017).
Pose-estimation networks that use deep learning can be trained to predict the spatial location of key body points in an image or video frame, and hence make physical markers placed on key body points obsolete. Pose-estimation networks enable repeated pose assessment on a large number of animals, which is needed to achieve accurate breeding values. Pose-estimation methods that use deep learning (Lecun et al., 2015) can learn the representation of key body points from annotated training data. In brief, these pose-estimation methods based on deep learning consists of two parts, a feature extractor that extracts visual features from a video image (frame), and a predictor that uses the output of the feature extractor to predict the body part and its location in the frame (Mathis et al., 2020). In part, the success of a supervised deep learning model depends on the availability of annotated data to learn these representations (Sun et al., 2017).
In the human domain, markerless pose estimation has been an active field of research for many years (e.g., Toshev and Szegedy, 2014; Insafutdinov et al., 2016; Sun et al., 2019; Cheng et al., 2020) and large datasets have been collected over the years [e.g., MPII (Andriluka et al., 2014), COCO (Lin et al., 2014)]. Animal pose estimation has been investigated in more recent studies (e.g., Mathis et al., 2018; Graving et al., 2019; Pereira et al., 2019), but large datasets remain scarce. One dataset (Cao et al., 2019) is publicly available, however, it is smaller than the human pose-estimation datasets and does not include broilers or turkeys. The creation of large datasets is difficult; large-scale animal data collection is not always easy, and data annotation is laborious and time-consuming. Therefore, efforts should be made to investigate methods that could permit deep-learning-based pose-estimation networks to work with limited data, and with that reduce annotation needs.
One method to work with limited data could be the use of data from different sources, like different species. Only a few studies have investigated the use of pose data from one or multiple species on another species (Sanakoyeu et al., 2020; Mathis et al., 2021). In Sanakoyeu et al. (2020), a chimpanzee pose-estimation network was trained on chimpanzee pseudo-labels originating from a network trained on data of humans and other species (bear, dog, elephant, cat, horse, cow, bird, sheep, zebra, giraffe, and mouse). Pseudo-labels are labels that are predicted by a model and not the result of manual annotation. In Mathis et al. (2021), a part of the research focused on the generalization of a pose-estimation network across species (horse, dog, sheep, cat, and cow). The pose-estimation network was trained on one or all other animal species whilst withholding either sheep or cow as test data. In both Mathis et al. (2021) and Sanakoyeu et al. (2020), despite differences in approach, pre-training with multiple species or training with multiple species resulted in better performance on the unseen species than when pre-training or training with one species. However, it is unclear whether the improved performance stems from a larger data availability or the multi-species data since no notion of dataset size was given. Furthermore, the investigated species were visually distinct, this might have affected the performance of the networks.
The objective of this study is to investigate the across-species performance of an animal pose-estimation network trained on broilers and tested on turkeys, and vice versa. Furthermore, since the interest is in working with limited data, the performance of an animal pose-estimation network trained on a multi-species training dataset (turkeys and broilers) will also be investigated. A multi-species dataset could potentially reduce annotation needs in both species without a negative effect on performance.
Materials and Methods
Data Collection
The data used in this research were collected in two different trials, one for turkeys and one for broilers. The data was not specifically collected for this study, but representative of the situation in practice. In both cases, the data collection will be presented separately though with a similar structure for easier comparison.
Turkeys
Data were collected on 83 male breeder turkeys at 20 weeks of age. This is approximately the slaughter age for commercial turkeys (Wood, 2009). The animals were subjected to a standard walkway test applied in the turkey breeding program of Hybrid Turkeys (Hendrix Genetics, Boxmeer, The Netherlands). The birds were stimulated to walk along a corridor (Width: ~1.5 m, Length: ~6 m) within the barn. Video recordings (RGB) were made from behind with an Intel® RealSense™ Depth Camera D415 (Intel Corporation, Santa Clara, United States; Resolution: 1,280 × 720, Frame Rate: 30). The camera was set up on a small tripod on a bucket to get a clear view of the legs of the birds. The camera was parallel to the ground and in the center of the walkway. A person trailed behind the birds to stimulate walking, and if needed waving their hand or tapping on the back of the bird. During the trial, the birds were equipped with three IMUs, one around the neck, the other two just above the hock. The IMU data was not used in this study but the IMUs were visible in the videos. Other birds were visible through wire-mesh fencing. The videos were cropped to a size of 600 × 720 to reduce the visibility of other turkeys through the wire-mesh fencing. The birds were housed under typical commercial circumstances.
Broilers
Data were collected on 47 conventional broilers at 37 days of age. The broilers were in the finishing stage and nearing the age of slaughter age at 41 days (Van Horne, 2020). The birds were stimulated to walk along a corridor (width: ~0.4 m, length: ~3 m) within the pen. Video recordings (RGB) were made from behind with the same Intel® RealSense™ Depth Camera D415 as used in the turkey experiment. The camera was set up in a fixed position on a metal rig attached to the front panel of the runway to get a clear view of the legs of the birds from behind. The camera was parallel to the ground and in the center of the walkway. The birds were stimulated to walk with a black screen made of wire netting on a stick. Other birds were not visible due to non-transparent side panels. The videos were not cropped since other broilers were not visible. The birds were housed in an experimental facility with a low stocking density (25 on 6 m2) but with standard light and feeding regime.
Frame Extraction and Annotation
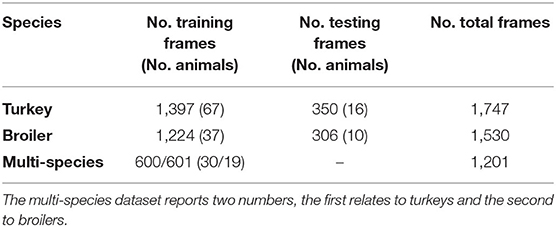
The toolbox of DeepLabCut 2.0 (version 2.1.8.2; Nath et al., 2019) was used to extract and annotate the frames from the collected RGB-videos (Table 1). For the turkeys, 20 frames per video/turkey were manually extracted to ensure no other animals were visible within the walkway and to exclude frames with human–animal interaction. For two turkeys, 50 frames were extracted. These two turkeys were part of our initial trial with DeepLabCut and hence had more annotated frames available. For the broilers, 40 frames per video/broiler were extracted, randomly sampled from a uniform distribution across time. The number of frames per broiler was roughly double the number of frames per turkey because the number of available broiler videos was roughly half that of the number of available turkey videos.
In principle, eight keypoints were annotated in each frame: head, neck, left knee, left hock, left foot, right knee, right hock, right foot (Figure 1). However, in some frames not all keypoints were visible (e.g., rump obscuring the head because the bird put its head down), these frames were retained, but the occluded keypoint was not annotated. The annotations are visually estimated locations founded on morphological knowledge, but can deviate from ground truth, particularly for keypoints obscured by plumage. The head was annotated at the top, the neck at the base, the knees at the estimated location of the knee, the hocks at the transition of the feathers into scales, and the feet approximately at the height of the first toe in the middle. The annotated data consisted of the x and y coordinates of the visible keypoints within the frames.
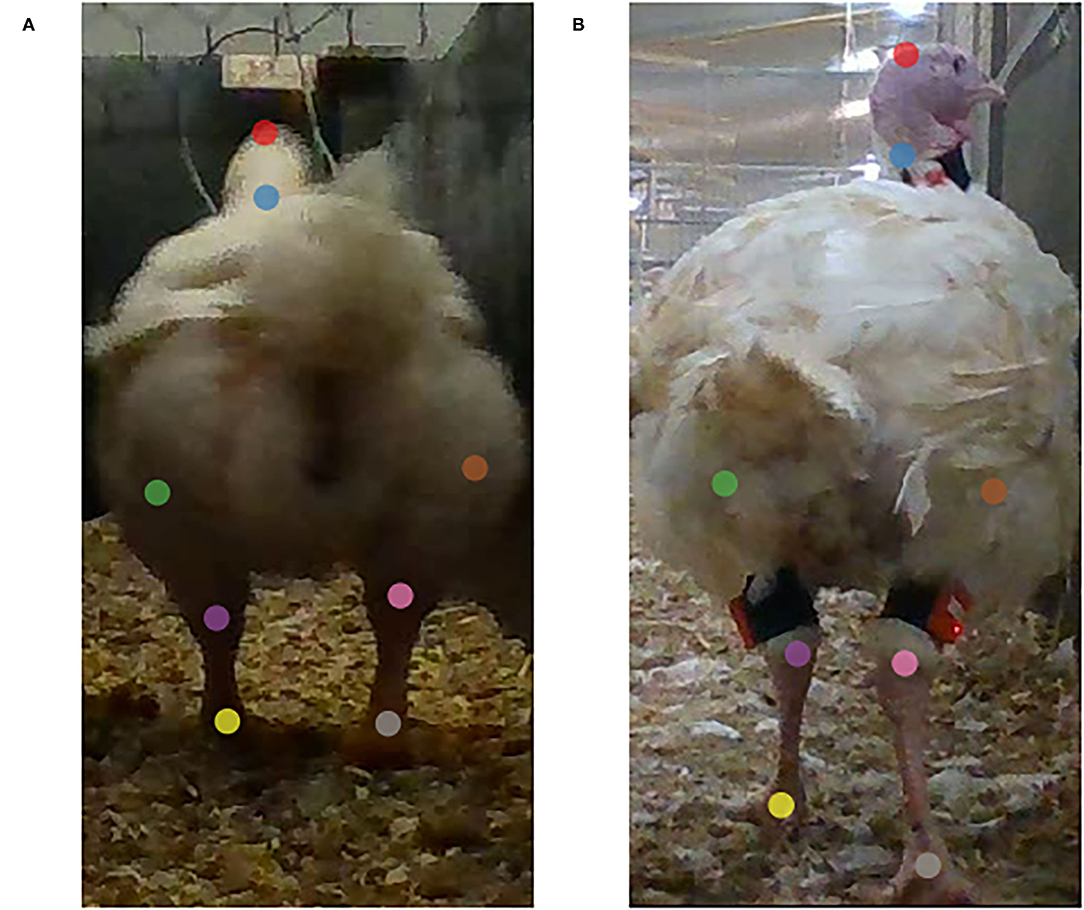
Figure 1. Example of broiler (A) and turkey (B) annotations. The keypoints are head (red), neck (blue), left knee (green), left hock (purple), left foot (yellow), right knee (brown), left hock (pink), and left foot (gray). Images are cropped.
Extracted frames with no animal in view or no visible keypoints (i.e., animal too close to the camera) were not annotated and subsequently removed. This only occurred in broiler frames, due to random frame extraction for the broilers vs. the manual frame extraction for the turkeys. Altogether, a total of 350 broiler frames were removed. There was no threshold on the minimal number of keypoints within a frame. In total, 3,277 frames were annotated by one annotator, consisting of 1,747 turkey frames and 1,530 broiler frames. The number of frames differed per animal (Table 1).
Datasets for Training and Testing
Five datasets were created from the annotated frames to train and test pose-estimation networks: two turkey datasets, two broiler datasets, and one multi-species training (turkey and broiler) dataset (Table 2). The single-species datasets were created by splitting the total number of frames in a training and test set (80 and 20%, respectively). Animals in the test set did not occur in the training set. Most animals in the test set were randomly selected, some were selected to get a proper 80/20-split since the number of frames differed per animal. The remainder of the frames made up the training data. The multi-species dataset was a combination of turkeys and broilers training frames. Most animals in the multi-species dataset were randomly selected from the animals in the turkey and broiler training set, some were selected to get the correct total number of frames. The five datasets thus consisted of three training datasets (turkey, broiler, multi-species) and two test datasets (turkey and broiler). An overview of the datasets is provided in Table 2.
Pose-Estimation
DeepLabCut is an open-source deep-learning-based pose-estimation tool (Mathis et al., 2018; Nath et al., 2019). In DeepLabCut, the feature detector from DeeperCut (Insafutdinov et al., 2016) is followed by deconvolutional layers to produce a score-map and a location refinement field for each keypoint. The score-map encodes the location probabilities of the keypoints (Figure 2). The location refinement field predicts an offset to counteract the effect of the down sampled score-map. The feature detector is a variant of deep residual neural networks (ResNet-50; He et al., 2016) pre-trained on ImageNet—a large-scale dataset for object recognition (Deng et al., 2009). The pre-trained network was fine-tuned for our task. This fine-tuning improves performance, reduces computational time, and reduces data requirements (Yosinski et al., 2014). During fine-tuning, the weights of the pre-trained network are iteratively adjusted on the training data of our task to ensure that the network returns high probabilities for the annotated keypoint locations (Mathis et al., 2018). DeepLabCut returns the location with the highest likelihood (θi) for each predicted keypoint in each frame (Figure 2).

Figure 2. Example of a broiler score-map. The score-map encodes the location probabilities of the keypoints.
Analyses
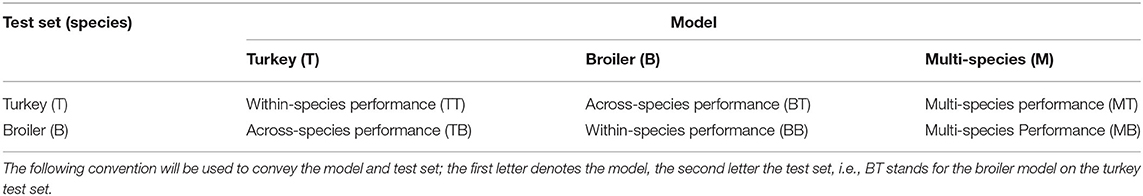
DeepLabCut (core, version 2.1.8.1; Mathis et al., 2018) was used to train three networks, one for each training dataset [turkey (T), broiler (B), and multi-species (M)]. All three networks were tested on both test datasets (turkey and broiler), thus within-species and across-species (Table 2). The model and test set will be indicated with the following convention; the first letter denotes the model, and the second letter the test set, i.e., MT stands for multi-species model on turkey test set and BB stands for broiler model on broiler test set.
All three networks were trained with default parameters for 1.03 million iterations (default). The number of epochs—the number times the entire dataset is presented through the network—differed between networks due to different training set sizes (turkey: 737 epochs; broiler: 841 epochs; multi-species: 858 epochs).
In Table 2, a testing scheme is presented. The testing scheme shows within-species (TT and BB), across-species (TB and BT) and multi-species model (MT and MB) testing. The within-species test established the performance of the networks on the species on which the model was trained. The across-species test was used to assess a network's performance across species, i.e., on the species on which the model was not trained. The multi-species model was tested on both test sets to assess the performance of a network trained with a combination of species and fewer annotations per species.
Evaluation Metrics
The performance of the models was evaluated with the raw pixel error, the normalized pixel error, and the percentage of keypoints remaining (PKR). The raw pixel error and normalized pixel error were calculated for all keypoints or keypoints with a likelihood higher or equal to 0.6 (default in DeepLabCut).
The raw pixel error was expressed as the Euclidean distance between the x and y coordinates of the model predictions and the human annotator.
Where dij is the Euclidean distance between the predicted location of keypoint i, , and its annotated location, (xi, yi), in frame j.
The average Euclidean distance was calculated per keypoint over all frames.
Where is the average Euclidean distance of keypoint i. N is the total number of frames, and N′ is the number of frames in which keypoint i was annotated, thus visible.
The overall average Euclidean distance was calculated over all keypoints over all frames.
Where is the overall average Euclidean distance, is the set of all valid annotations of all keypoints i in all frames j.
Since the animal is moving away from the camera, the size of the animal in relation to the frame changes, i.e., the animal becomes smaller. The normalized pixel error corrects the raw pixel error for the size of the animal in the frame, i.e., a pixel error of five pixels when the animal is near the camera is better than a pixel error of five pixels when the animal is further from the camera. The raw pixel errors were normalized by the square root of the bounding box area of the legs, as head and neck keypoints were not always visible. The bounding box was constructed from the annotated keypoints to ensure that the normalization of the raw pixel error was independent of the predictions. The square root of the bounding box area penalized the pixel errors less for large bounding boxes than for small bounding boxes.
The normalized pixel error was calculated as follows:
Where dij is the raw pixel error as in Equation (1), is a set of annotated leg keypoint coordinates, (xi, yi), in frame j. Leg keypoints consist of the knees, the hocks, and the feet. The normalized pixel error was reported either as the average normalized error per keypoint as in Equation (2) or as the overall average normalized error as in Equation (3) with dij substituted with Normdij.
The PKR is the percentage of keypoints with a likelihood higher or equal to 0.6 over the total keypoints with a Euclidean distance. Only annotated keypoints have a Euclidean distance (see also Equation 1). The PKR is a proxy for the confidence of the model. The PKR should always be considered in unison with the pixel error, a model with a high PKR and a low pixel error is confidently right.
Results
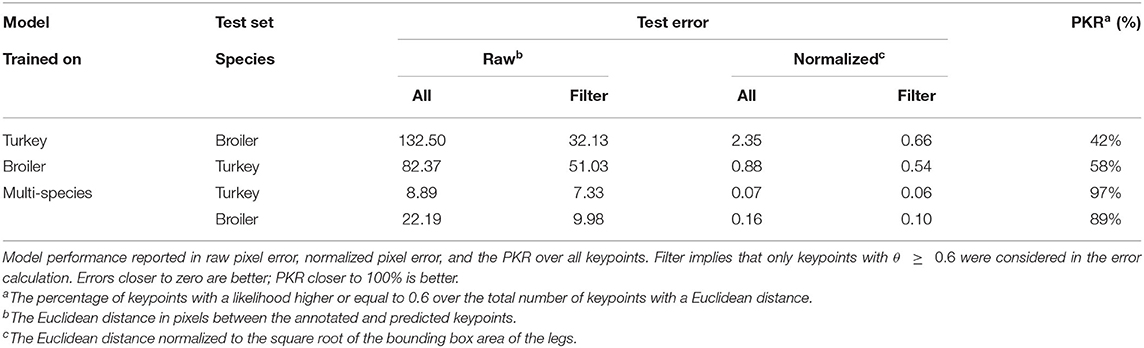
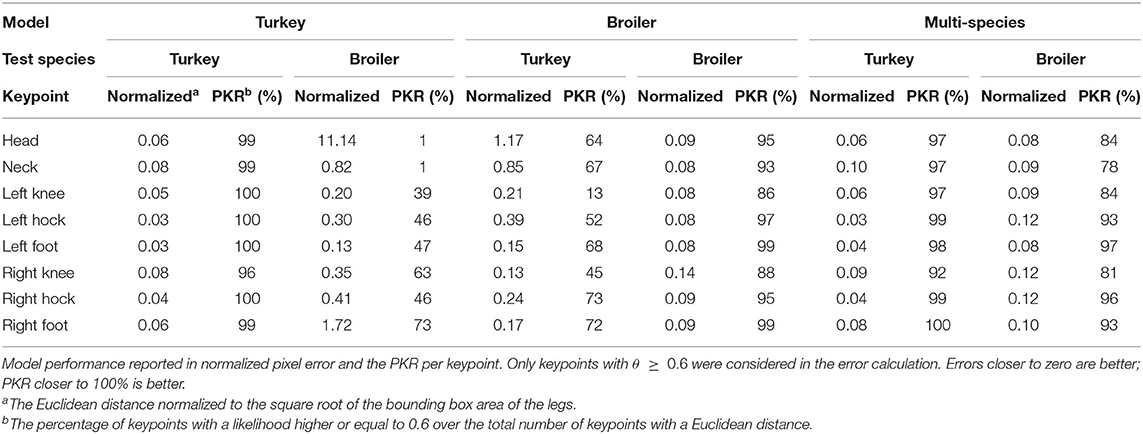
The models were used to investigate the across-species performance of animal pose-estimation networks and the performance of an animal pose-estimation network trained on multi-species data. The models were tested according to the testing scheme in Table 3. The performances of all models over all keypoints are shown in Tables 4, 5.
Comparison Between Within-Species, Across-Species, and Multi-Species
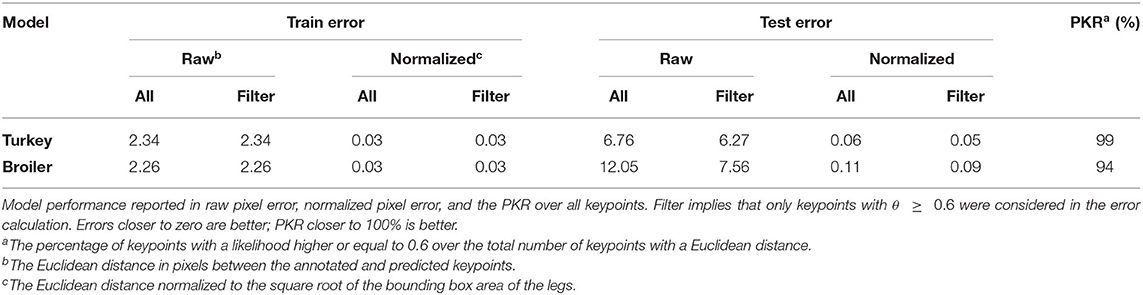
On all evaluation metrics calculated overall keypoints, the within-species models (TT, BB) outperformed the multi-species model (MT, MB) and the models applied across species (TB, BT) (Tables 4, 5). The within-species models had lower raw pixel errors, lower normalized pixel errors, and higher PKRs than the multi-species model and the models applied across-species. Compared to the within-species models, the multi-species model had slightly higher normalized errors (+0.01). However, the errors across-species were considerably higher (+0.57; +0.49) than they were for the within-species models.
Performance varied per keypoint, not only within models but also between models (Table 6). In general, the head, neck and knee keypoints were predicted with the highest errors. Across-species, the models always performed worse than the within-species counterpart and the multi-species model. On the broiler test set, the multi-species model outperformed the broiler model for the head and right knee keypoint, although this did coincide with a lower PKR. The turkey model had either a similar or better performance than the multi-species model on the turkey test set but the multi-species model did generally have a lower PKR.
Within-Species
On the training dataset, both within-species models (TT, BB) showed comparable raw pixel errors and normalized pixel errors (Table 4). The turkey model (TT) had a lower raw and normalized pixel error and higher PKR than the broiler model (BB). The turkey model had the lowest keypoints error for the left hock and left foot and the highest error for the right knee keypoint (Table 6). The right knee keypoint error was 0.03 higher than the left knee keypoint error. The leg keypoint errors of the broiler model were rather consistent within each leg, except for the right knee keypoint.
Multi-Species
Multi-species model performance was different between species (MT, MB; Table 4). The multi-species model performed better on the turkey test set (MT) than it did on the broiler test set (MB). The multi-species model on the turkey test set had the highest error for the neck keypoint and the lowest error for the left hock keypoint (Table 6). On the broiler test set, the multi-species model had the highest errors for the hocks and right knee keypoints, and the lowest error for the head keypoint.
Across-Species
Across species, the turkey and broiler model had high errors (TB, BT; Table 5). The turkey model on the broiler test set had the highest error for the head keypoint, whereas the left foot keypoint had the lowest error (Table 6). The broiler model on turkey test set also had the highest error for the head keypoint and lowest error for the left foot keypoint.
Discussion
In this study, the across-species performance of animal pose-estimation networks and the performance of an animal pose-estimation network trained on multi-species data (turkeys and broilers) were investigated. The results showed that within-species the models had the best performance, followed by the multi-species model, and across-species the models had the worst performance, as illustrated by the raw pixel errors, normalized pixel errors, and PKRs. However, the multi-species model outperformed the broiler model on the broiler test set for the head, left foot, and right knee keypoints, though with a lower PKR.
Data Availability and Model Performance
The turkey model outperformed the broiler model on the within species test set (Table 4), even though both models had approximately comparable raw pixel errors and normalized pixel errors on the training dataset. For the turkeys, the training set was slightly larger (n = 1,397) than the broiler training set (n = 1,224), which might explain the difference in performance. However, the turkey test set was likely less challenging, as the difference between the unfiltered and filtered error was smaller for the turkey model than it was for the broiler model. The difference in difficulty can partly be explained by the difference in frame extraction. The broiler dataset consisted of frames that were randomly sampled from a uniform distribution across time, whereas the turkey dataset consisted of consecutive frames. The temporal correlation between the frames may explain why the turkey test set was less challenging.
Overall, the multi-species model had higher errors and a lower PKR than the single-species models. Yet, compared to the within-species models, the multi-species model had less than half the number of annotated frames of the tested species. Interestingly, the multi-species model performed better or similar for certain keypoints compared to the single-species models, but with less confidence, hence a lower PKR. The multi-species model performance suggests that data from the other species was useful to improve performance for certain keypoints but did lower the PKR. The lower PKR is more apparent on the broiler test set but also noticeable on the turkey test set. The lower PKR may be caused by an interplay between the inclusion of other species training data and a lower variability within the species-specific training data.
The pose-estimation networks applied across species had no data available on the target species and could still estimate keypoints. Those keypoint estimates appear to be relatively informed as indicated by the normalized errors. This suggests that, in the case of comparable species, with an existing model and limited availability of data on the new species, the existing model could be fine-tuned on limited data of the target species. The performance of the pose-estimation models confirmed that the success of a supervised deep learning model depends on the availability of data, as was noted by Sun et al. (2017).
Across-species, the head and neck showed high normalized pixel errors for both turkeys and broilers. Across-species pose-estimation is influenced by differences in appearance of the animals and the differences in environment. There are inherent differences in appearance between turkeys and broilers, especially concerning the head and neck. A turkey head is featherless and has a light-blue tint and a broiler head is feathered and white. In our case, it appears that DeepLabCut was dependent on the color of the keypoints. The broiler model tended to predict the turkey head in the white overhead lights, on workers' white boots, and turkeys at the end of the walkway. These locations were relatively far away from the bird, as indicated by the pixel error. A model that uses spatial information of other keypoints within a frame could notice that these predicted keypoints are too far off and search for the second-best location closer to the other keypoints. This suggests that single animal DeepLabCut could benefit from the use of spatial information of other keypoints within a frame, as was also noted by Labuguen et al. (2021).
Data Collection
In this study, the data was collected in two different trials, one for turkeys and one for broilers but neither specific for this study. Recording both species in the same setting under the same conditions may have been better for assessing model performance between the two species, but can only be done in an experimental setting, which often poorly translates to practical implementation. The datasets used here were representative of the situation in practice for poultry breeding programs. In the end, the models will have to work in less regulated environments, i.e., barns and pens, to be of use.
In the turkey trial, multiple sensors collected data to assess the gait of the animals. The trial did not only involve a video camera, but the animals were also equipped with IMUs, and there was a force plate hidden underneath the bedding. The IMUs were attached to both legs and the neck, and hence they were visible in the turkey video frames. The presence of the IMUs was likely picked up by the pose-estimation network, as the hocks often had the lowest normalized pixel error, and highest PKR of all keypoints within a turkey leg. Likewise, when the broiler model was tested on the turkey test set it tended to predict the hocks at the transition of the Velcro strap of the IMU to the feathers, instead of the transition from scales into feathers. The presence of external sensors seems to have influenced the performance of the pose-estimation networks on the turkey test set.
The turkey trial was conducted during a standard walkway test applied in the turkey breeding program of Hybrid Turkeys (Hendrix Genetics, Boxmeer, The Netherlands), and therefore, representative of a practical gait scoring situation. The turkeys were stimulated to walk by a worker causing occlusions in the frames. However, occlusions could also occur because of another bird in queue, while the bird of interest was still walking. In the turkey dataset, only frames without occlusion by a worker or other bird were included. These occlusions limit the amount of usable data available for gait and pose estimations. The occlusions did not hinder the human expert who can move around freely, while the camera is in a fixed position. In an ideal situation, each animal is walked one-by-one for the full extent of the walkway, as was done with the broilers. This will not only make the videos more usable but also allow for a better sampling of the frames to train a network.
Annotation
During the annotation process, not all keypoints could be annotated as accurately. For both turkeys and broilers, the knees were annotated at the estimated location as the knees of the birds cannot be observed directly. The uncertainty in labeling, and thereby the variability in labeling, declined when the animal was further away from the camera since the likely knee area simply declined, but annotator uncertainty was still present. The larger likely knee area when the animal was near the camera, coupled with the annotator uncertainty is likely to increase the raw pixel errors. The annotator uncertainty probably increased the variability of the knee keypoint annotations which would have a negative effect on the PKR, as the network would have more trouble learning the knee keypoint. The annotator uncertainty becomes evident when we look at the normalized pixel error and the PKR of the turkey and broiler model applied within species. The knees had the highest normalized pixel error and/or lowest PKR of the keypoints within each leg. Ideally, the normalized pixel error of the knees reflected the decline of the likely knee area by being equal to the normalized pixel error of the other keypoints within the leg. However, the normalized pixel error of the knee keypoints was only equal to the normalized pixel error of the other keypoins within the left leg of the broilers, in all other cases, it was higher, showing that labeling uncertainty was still present.
Prospects
This study provides insight into the across-species performance of animal pose-estimation networks and the performance of an animal pose-estimation network trained on multi-species data. Accurate pose-estimation networks enable automated estimation of key body points in an images or video frames, which are a prerequisite to use camera's for objective assessment of poses and gaits, hence within species trained models would perform best, if sufficient annotated data is available on the species. Within-species models will provide more accurate keypoints from which more accurate spatiotemporal (e.g., step time and speed) and kinematic (e.g., joint angles) gait and pose parameters can be estimated. In case of limited data availability, a multi-species model could potentially be considered for pose assessment without a large impact on performance if the used species are comparable. The across-species keypoint estimates may not be precise enough for accurate gait and pose assessments, but still appear to be relatively informed as indicated by the normalized errors. A pose estimation network may not be directly applicable across species, but the network could serve as a pre-trained network that can be fine-tuned on the target species if there is limited available data. An alternative could be the use of Generative Adversarial Neural networks (GANs; Zhu et al., 2017). However, recent GANs appear to work better to change coat color than to change a dog into a cat (Cao et al., 2019). Furthermore, if the species change is successful, the accuracy of the converted keypoint labels could be negatively impacted (Cao et al., 2019).
Conclusion
In this study, the across-species performance of animal pose-estimation networks and the performance of an animal pose-estimation network trained on multi-species data (turkeys and broilers) were investigated. Across species, keypoint predictions resulted in high errors in low to moderate PKRs and are unlikely to be of direct use for pose and gait assessments. The multi-species model had slightly higher errors with a lower PKR than the within-species models but had less than half the number of annotated frames available from each species. The within-species model had the overall best performance. The within-species models will provide more accurate keypoints from which more accurate spatiotemporal and kinematic—geometric and time-dependent aspects of motion—gait and pose parameters can be estimated. A multi-species model could potentially reduce annotation needs without a large impact on performance on pose assessment, however, with the recommendation to only be used if the species are comparable. Future studies should investigate the actual accuracy needed for pose and gait assessments and estimate genetic parameters for the new phenotypes before pose-estimation networks can be applied in practice.
Data Availability Statement
The turkey dataset analyzed for this study is not publicly available as it is the intellectual property of Hendrix Genetics. Requests to access the dataset should be directed to Bram Visser, YnJhbS52aXNzZXJAaGVuZHJpeGdlbmV0aWNzLmNvbQ==. The broiler dataset analyzed for this study is available upon reasonable request from Wageningen Livestock Research. Requests to access the dataset should be directed to Aniek C. Bouwman, YW5pZWsuYm91d21hbkB3dXIubmw=.
Ethics Statement
The Animal Welfare Body of Wageningen Research decided ethical review was not necessary because the turkeys were not isolated, semi-familiar with the corridor, the IMUs low in weight (1% of body weight), and the IMUs were attached for not longer than one hour. The Animal Welfare Body of Wageningen University noted that the broiler study did not constitute an animal experiment under Dutch law. The experimental procedures described in the protocol of the broiler study would cause less pain or distress than the insertion of a needle under good veterinary practice.
Author Contributions
JD, AB, GK, and RV contributed to the conceptualization of the study. JD, AB, and JE were involved with broiler data collection. JD and TS performed annotation and analysis. JD wrote the first draft of the manuscript. JD, AB, EE, JE, GK, and RV reviewed and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financially supported by the Dutch Ministry of Economic Affairs (TKI Agri and Food project 16022) and the Breed4Food partners Cobb Europe, CRV, Hendrix Genetics and Topigs Norsvin.
Conflict of Interest
The authors declare that this study received funding from Cobb Europe, CRV, Hendrix Genetics and Topigs Norsvin. The funders had the following involvement in the study: All funders were involved in the study design. Hendrix Genetics was involved in data collection. None of the funders was involved in the analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the Hybrid Turkeys team (Kitchener, Canada) for the collection of the turkey data. We thank Henk Gunnink and Stephanie Melis (Animal Health and Welfare, Wageningen University and Research) for their help with broiler data collection. We would also like to thank Ard Nieuwenhuizen (Agrosystems Research, Wageningen University and Research) for his work on the turkey data and introducing us to DeepLabCut. The use of the HPC cluster has been made possible by CAT-AgroFood (Shared Research Facilities Wageningen UR).
References
Abourachid, A (1991). Comparative gait analysis of two strains of turkey, meleagris gallopavo. Br. Poult. Sci. 32, 271–277. doi: 10.1080/00071669108417350
Abourachid, A (2001). Kinematic parameters of terrestrial locomotion in cursorial (ratites), swimming (ducks), and striding birds (quail and guinea fowl). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 131, 113–119. doi: 10.1016/S1095-6433(01)00471-8
Andriluka, M., Pishchulin, L., Gehler, P., and Schiele, B. (2014). “2D human pose estimation: new benchmark and state of the art analysis,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (Columbus, OH), 3686–3693. doi: 10.1109/CVPR.2014.471
Bassler, A. W., Arnould, C., Butterworth, A., Colin, L., De Jong, I. C., Ferrante, V., et al. (2013). Potential risk factors associated with contact dermatitis, lameness, negative emotional state, and fear of humans in broiler chicken flocks. Poult. Sci. 92, 2811–2826. doi: 10.3382/ps.2013-03208
Bouwman, A., Savchuk, A., Abbaspourghomi, A., and Visser, B. (2020). Automated Step detection in inertial measurement unit data from turkeys. Front. Genet. 11:207. doi: 10.3389/fgene.2020.00207
Bradshaw, R. H., Kirkden, R. D., and Broom, D. M. (2002). A review of the aetiology and pathology of leg weakness in broilers in relation to their welfare. Avian Poult. Biol. Rev. 13, 45–104. doi: 10.3184/147020602783698421
Cao, J., Tang, H., Fang, H. S., Shen, X., Lu, C., and Tai, Y. W. (2019). “Cross-domain adaptation for animal pose estimation,” in Proceedings of the IEEE/CVF International Conference on Computer Vision Vol. 1 (Seoul), 9497–9506. doi: 10.1109/ICCV.2019.00959
Caplen, G., Hothersall, B., Murrell, J. C., Nicol, C. J., Waterman-Pearson, A. E., Weeks, C. A., et al. (2012). Kinematic analysis quantifies gait abnormalities associated with lameness in broiler chickens and identifies evolutionary gait differences. PLoS ONE 7:e40800. doi: 10.1371/journal.pone.0040800
Cheng, B., Xiao, B., Wang, J., Shi, H., Huang, T. S., and Zhang, L. (2020). “Higherhrnet: scale-aware representation learning for bottom-up human pose estimation,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (Seattle, WA), 5386–5395.
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., and Fei-Fei, L. (2009). ImageNet: a large-scale hierarchical image database. IEEE Conf. Comp. Vis. Patt. Recogn. (Miami, FL), 248–255. doi: 10.1109/cvpr.2009.5206848
Erasmus, M. A (2018). Welfare issues in turkey production. in Advances in Poultry Welfare, ed J. A. Mench (Duxford: Woodhead Publishing), 263–291. doi: 10.1016/B978-0-08-100915-4.00013-0
Garner, J. P., Falcone, C., Wakenell, P., Martin, M., and Mench, J. A. (2002). Reliability and validity of a modified gait scoring system and its use in assessing tibial dyschondroplasia in broilers. Br. Poult. Sci. 43, 355–363. doi: 10.1080/00071660120103620
Graving, J. M., Chae, D., Naik, H., Li, L., Koger, B., Costelloe, B. R., et al. (2019). Deepposekit, a software toolkit for fast and robust animal pose estimation using deep learning. Elife 8, 1–42. doi: 10.7554/eLife.47994
He, K., Zhang, X., Ren, S., and Sun, J. (2016). “Deep residual learning for image recognition,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (Las Vegas, NV), 770–778.
Insafutdinov, E., Pishchulin, L., Andres, B., Andriluka, M., and Schiele, B. (2016). “DeeperCut: a deeper, stronger, and faster multi-person pose estimation model,” in European Conference on Computer Vision (Amsterdam), 34–50.
Kapell, D. N. R. G., Hocking, P. M., Glover, P. K., Kremer, V. D., and Avendaño, S. (2017). Genetic basis of leg health and its relationship with body weight in purebred turkey lines. Poult. Sci. 96, 1553–1562. doi: 10.3382/ps/pew479
Kestin, S. C., Knowles, T. G., Tinch, A. E., and Gregory, N. G. (1992). Prevalence of leg weakness in broiler chickens and its relationship with genotype. Vet. Rec. 131, 190–194. doi: 10.1136/vr.131.9.190
Kittelsen, K. E., David, B., Moe, R. O., Poulsen, H. D., Young, J. F., and Granquist, E. G. (2017). Associations among gait score, production data, abattoir registrations, and postmortem tibia measurements in broiler chickens. Poult. Sci. 96, 1033–1040. doi: 10.3382/ps/pew433
Kremer, J. A., Robison, C. I., and Karcher, D. M. (2018). Growth dependent changes in pressure sensing walkway data for Turkeys. Front. Vet. Sci. 5:241. doi: 10.3389/fvets.2018.00241
Labuguen, R., Matsumoto, J., Negrete, S. B., Nishimaru, H., Nishijo, H., Takada, M., et al. (2021). MacaquePose: a novel “in the wild” macaque monkey pose dataset for markerless motion capture. Front. Behav. Neurosci. 14:581154. doi: 10.3389/fnbeh.2020.581154
Lecun, Y., Bengio, Y., and Hinton, G. (2015). Deep learning. Nature 521, 436–444. doi: 10.1038/nature14539
Lin, T. Y., Maire, M., Belongie, S., Hays, J., Perona, P., Ramanan, D., et al. (2014). “Microsoft COCO: common objects in context,” in European Conference on Computer Vision (Zurich), 740–755. doi: 10.1007/978-3-319-10602-1_48
Malchow, J., Dudde, A., Berk, J., Krause, E. T., Sanders, O., Puppe, B., et al. (2019). Is the rotarod test an objective alternative to the gait score for evaluating walking ability in chickens? Anim. Welf. 28, 261–269. doi: 10.7120/109627286.28.3.261
Mathis, A., Mamidanna, P., Cury, K. M., Abe, T., Murthy, V. N., Mathis, M. W., et al. (2018). DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci. 21, 1281–1289. doi: 10.1038/s41593-018-0209-y
Mathis, A., Schneider, S., Lauer, J., and Mathis, M. W. (2020). A primer on motion capture with deep learning: principles, pitfalls, and perspectives. Neuron 108, 44–65. doi: 10.1016/j.neuron.2020.09.017
Mathis, A., Yüksekgönül, M., Rogers, B., Bethge, M., and Mathis, M. W. (2021). “Pretraining boosts out-of-domain robustness for pose estimation,” in Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (Waikoloa, HI), 1859–1868.
Nääs, I. D. A., de Lima Almeida Paz, I. C., Baracho, M. d. S., de Menezes, A. G., de Lima, K. A. O., et al. (2010). Assessing locomotion deficiency in broiler chicken. Sci. Agric. 67, 129–135. doi: 10.1590/S0103-90162010000200001
Nath, T., Mathis, A., Chen, A. C., Patel, A., Bethge, M., and Mathis, M. W. (2019). Using DeepLabCut for 3D markerless pose estimation across species and behaviors. Nat. Protoc. 14, 2152–2176. doi: 10.1038/s41596-019-0176-0
Oviedo-Rondón, E. O., Lascelles, B. D. X., Arellano, C., Mente, P. L., Eusebio-Balcazar, P., Grimes, J. L., et al. (2017). Gait parameters in four strains of turkeys and correlations with bone strength. Poult. Sci. 96, 1989–2005. doi: 10.3382/ps/pew502
Paxton, H., Daley, M. A., Corr, S. A., and Hutchinson, J. R. (2013). The gait dynamics of the modern broiler chicken: a cautionary tale of selective breeding. J. Exp. Biol. 216, 3237–3248. doi: 10.1242/jeb.080309
Pereira, T. D., Aldarondo, D. E., Willmore, L., Kislin, M., Wang, S. S. H., Murthy, M., et al. (2019). Fast animal pose estimation using deep neural networks. Nat. Methods 16, 117–125. doi: 10.1038/s41592-018-0234-5
Quinton, C. D., Wood, B. J., and Miller, S. P. (2011). Genetic analysis of survival and fitness in turkeys with multiple-trait animal models. Poult. Sci. 90, 2479–2486. doi: 10.3382/ps.2011-01604
Sanakoyeu, A., Khalidov, V., McCarthy, M. S., Vedaldi, A., and Neverova, N. (2020). “Transferring dense pose to proximal animal classes,” in Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition (Seattle, WA), 5233–5242. doi: 10.1109/CVPR42600.2020.00528
Scientific Committee on Animal Health and Animal Welfare (2000). The Welfare of Chickens Kept for Meat Production (Broilers). European Union Commission.
Sharafeldin, T. A., Mor, S. K., Bekele, A. Z., Verma, H., Noll, S. L., Goyal, S. M., et al. (2015). Experimentally induced lameness in turkeys inoculated with a newly emergent turkey reovirus. Vet. Res. 46, 1–7. doi: 10.1186/s13567-015-0144-9
Stevenson, R., Dalton, H. A., and Erasmus, M. (2019). Validity of micro-data loggers to determine walking activity of turkeys and effects on turkey gait. Front. Vet. Sci. 5:319. doi: 10.3389/fvets.2018.00319
Sullivan, T. W (1994). Skeletal problems in poultry: estimated annual cost and descriptions. Poult. Sci. 73, 879–882. doi: 10.3382/ps.0730879
Sun, C., Shrivastava, A., Singh, S., and Gupta, A. (2017). “Revisiting unreasonable effectiveness of data in deep learning era,” in Proceedings of the IEEE International Conference on Computer Vision (Venice), 843–852. doi: 10.1109/ICCV.2017.97
Sun, K., Xiao, B., Liu, D., and Wang, J. (2019). “Deep high-resolution representation learning for human pose estimation,” in Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition (Long Beach, CA), 5686–5696.
Toshev, A., and Szegedy, C. (2014). “DeepPose: human pose estimation via deep neural networks,” Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition (Columbus, OH), 1653–1660. doi: 10.1109/CVPR.2014.214
Van Horne, P. L. M (2020). Economics of Broiler Production Systems in the Netherlands: Economic Aspects Within the Greenwell Sustainability Assessment Model. Available online at: https://www.wur.nl/en/show/Report-Economics-of-broiler-production-systems-in-the-Netherlands.htm (accessed September 11, 2021).
van Staaveren, N., Leishman, E. M., Wood, B. J., Harlander-Matauschek, A., and Baes, C. F. (2020). Farmers' perceptions about health and welfare issues in turkey production. Front. Vet. Sci. 7:332. doi: 10.3389/fvets.2020.00332
Vermette, C., Schwean-Lardner, K., Gomis, S., Grahn, B. H., Crowe, T. G., and Classen, H. L. (2016). The impact of graded levels of day length on Turkey health and behavior to 18 weeks of age. Poult. Sci. 95, 1223–1237. doi: 10.3382/ps/pew078
Vestergaard, K. S., and Sanotra, G. S. (1999). Changes in the behaviour of broiler chickens. Vet. Rec. 144, 205–210.
Weeks, C. A., Danbury, T. D., Davies, H. C., Hunt, P., and Kestin, S. C. (2000). The behaviour of broiler chickens and its modification by lameness. Appl. Anim. Behav. Sci. 67, 111–125. doi: 10.1016/S0168-1591(99)00102-1
Wood, B. J (2009). Calculating economic values for turkeys using a deterministic production model. Can. J. Anim. Sci. 89, 201–213. doi: 10.4141/CJAS08105
Yosinski, J., Clune, J., Bengio, Y., and Lipson, H. (2014). How transferable are features in deep neural networks? Adv. Neural Inf. Process. Syst. (Montreal, QC), 4, 3320–3328. Available online at: https://proceedings.neurips.cc/paper/2014/file/375c71349b295fbe2dcdca9206f20a06-Paper.pdf
Keywords: broilers, computer vision, deep learning, gait, multi-species, pose-estimation, turkeys, within-species
Citation: Doornweerd JE, Kootstra G, Veerkamp RF, Ellen ED, van der Eijk JAJ, van de Straat T and Bouwman AC (2021) Across-Species Pose Estimation in Poultry Based on Images Using Deep Learning. Front. Anim. Sci. 2:791290. doi: 10.3389/fanim.2021.791290
Received: 08 October 2021; Accepted: 24 November 2021;
Published: 15 December 2021.
Edited by:
Dan Børge Jensen, University of Copenhagen, DenmarkReviewed by:
Madonna Benjamin, Michigan State University, United StatesE. Tobias Krause, Friedrich-Loeffler-Institute, Germany
Copyright © 2021 Doornweerd, Kootstra, Veerkamp, Ellen, van der Eijk, van de Straat and Bouwman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jan Erik Doornweerd, amFuZXJpay5kb29ybndlZXJkQHd1ci5ubA==
 Jan Erik Doornweerd
Jan Erik Doornweerd Gert Kootstra2
Gert Kootstra2 Roel F. Veerkamp
Roel F. Veerkamp Esther D. Ellen
Esther D. Ellen Jerine A. J. van der Eijk
Jerine A. J. van der Eijk Aniek C. Bouwman
Aniek C. Bouwman